Abstract
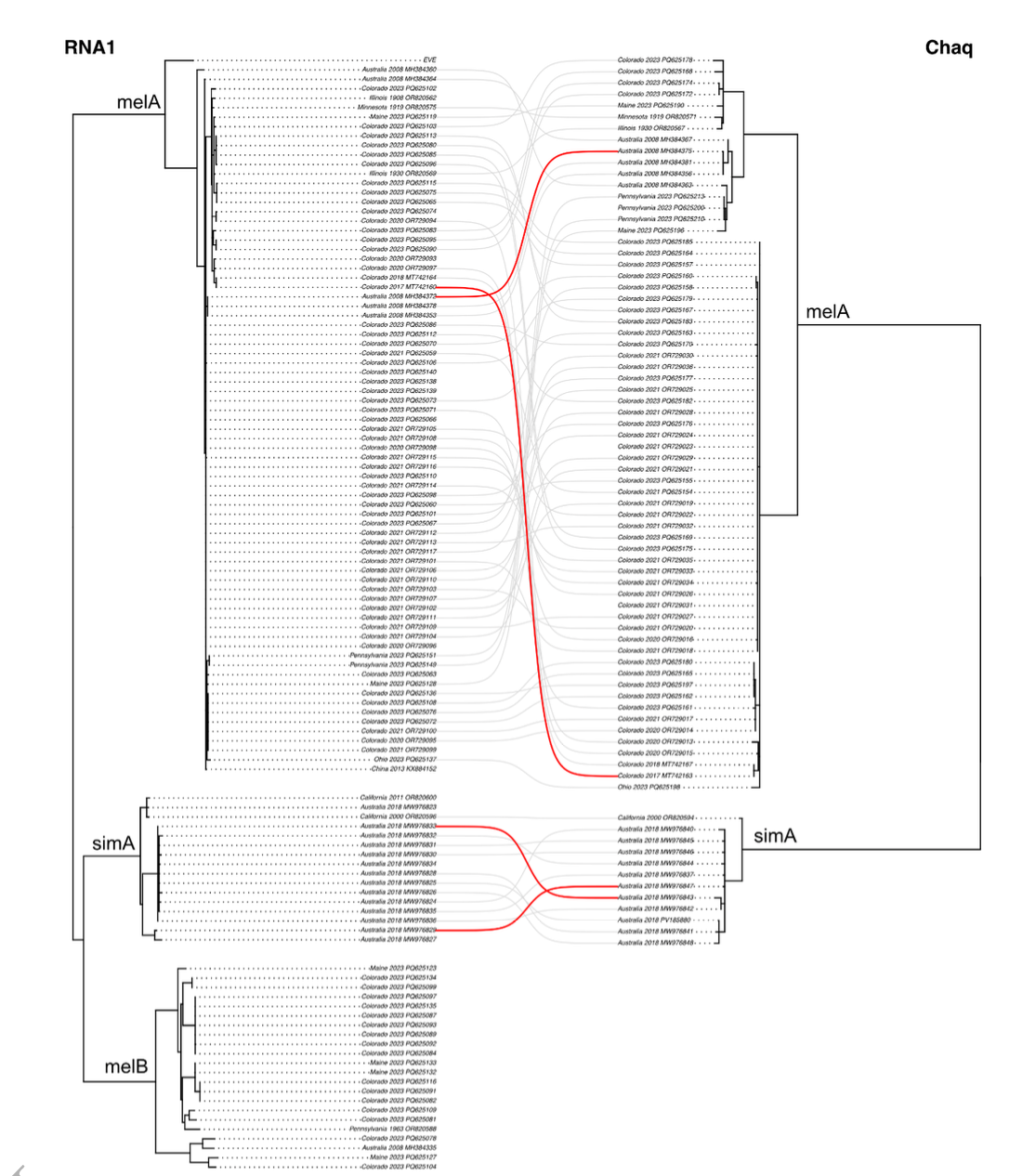
Galbut virus is a remarkably successful virus of the model organism Drosophila melanogaster. The ease of capturing galbut virus infected wild flies presents an opportunity to study persistent virus-host interactions. To better understand galbut virus diversity, evolution, and genotype-phenotype relationships, we screened 957 flies collected from 13 locations across the USA. Galbut virus was detected in every population and overall, 75% of flies tested positive. We selected 149 flies for shotgun RNA sequencing and recovered 368 coding-complete or near-complete sequences of galbut virus and its likely satellite, chaq virus. Galbut virus sequences clustered phylogenetically into three major clades. Two clades, termed melA and melB, comprised mainly viruses infecting D. melanogaster while the third, simA, included D. simulans-infecting viruses. Between-clade pairwise nucleotide identity was as low as 75%, but diversity within clades was low. Geography partly explained phylogenetic clustering: some sequences from the same location were identical or nearly identical while others were spread throughout trees. Several genotype-phenotype associations emerged, including higher average RNA levels in melA infections and an exclusive association of chaq virus with melA and simA infections. Coinfection was detected in 11% of samples and did not require complete sets of all three galbut virus segments. Coinfection is a prerequisite of reassortment, and there was evidence of reassortment involving all segments and chaq virus. However, reassortment did not occur between clades, despite coinfection, indicating that clades are reproductively isolated. Galbut virus RNA 3 sequences exhibited more involvement in coinfection, greater diversity, and stronger evidence of diversifying selection than the other segments, consistent with a possible role in host modulation. These findings corroborated the ecological success of galbut virus and reveal the need for experiments to uncover the mechanisms underlying reassortment incompatibility and clade-specific phenotypes.